Reseach projects
Cancer stem cells from triple negative breast cancer – novel therapies and therapeutics
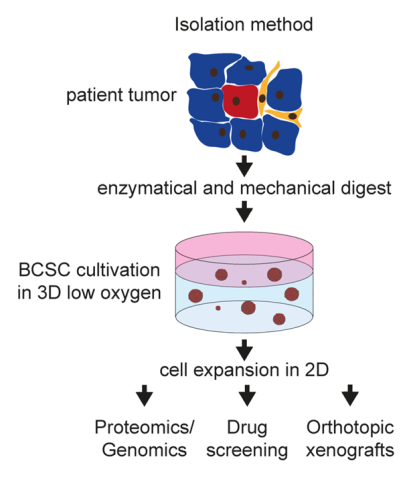
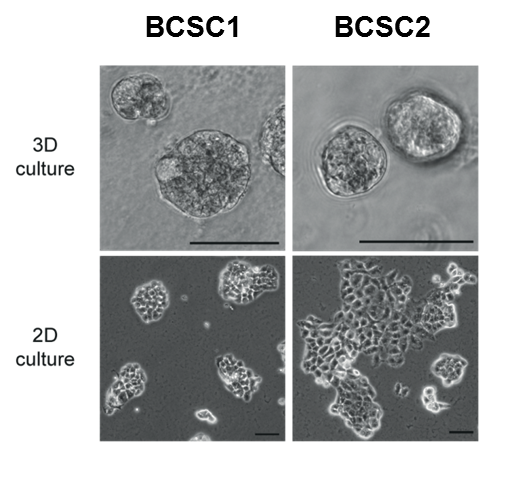
Triple negative breast cancer represents a highly aggressive subtype of breast cancer with 9000-14000 new cases in Germany each year (data from 2011-2017) and is characterized by a significant higher frequency of visceral as well as central nervous system metastases than the other subtypes. Due to this clinical behaviour TNBC shows a poor prognosis profile with significant lower recurrence free and overall survival rates, respectively. Breast cancer stem cells (BCSCs) are hypothesized to play a crucial role for the tumorbiological behavior of TNBC with the high metastatic potential as well as the observed metastatic pattern. We were able to isolate and characterize these cells (Fig. 1) in vitro and in vivo (Metzger and Stepputtis et al, 2017).
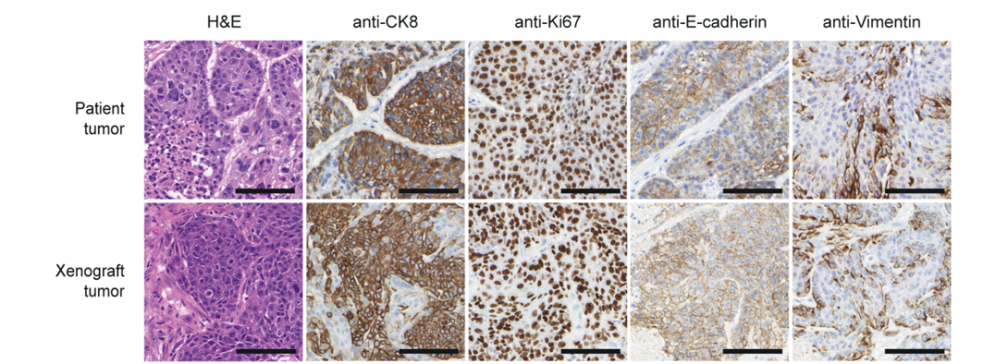
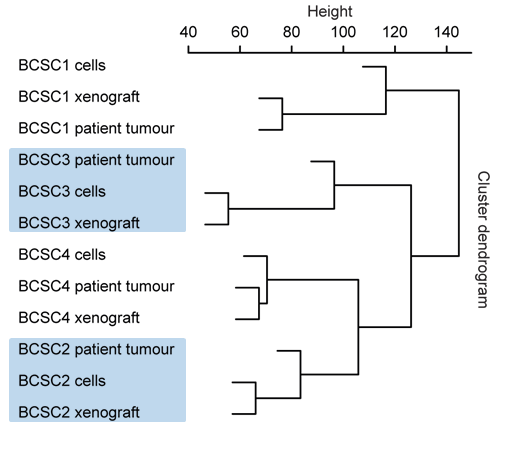
Fig. 2: BCSCs represent the host tumor in an orthotopic Xenograft immunohistochemically (left) and on RNA level (right).
BCSCs represent the patient tumor in vitro and in an orthotopic xenograft (Fig. 2) from as few as a 1000 cells making these cells the ideal basis for our multiple projects:
UCARE
Identification of key modulators of microRNA expression in breast cancers and their potential as prognostic and predictive biomarkers for innovative neoadjuvant therapeutic options
With breast cancer still affecting about one third of the female population of this planet in their lifetime, improvements to diagnosis and therapy are of the utmost importance. In conjunction with established methods for early detection, the treatment and, consequently, the quality of life and the survival of women with breast cancer could be significantly improved with the help of targeted individualized neoadjuvant chemotherapy. The advantage of this concept is that, at the molecular biological level, specific neoadjuvant chemotherapy with the best possible treatment success can be selected with the help of specific breast cancer-associated biomarkers. Due to the good results in this field on the one hand and the knowledge about the extraordinarily strong heterogeneity of breast cancer tumors, the search for further biomarkers is becoming increasingly important. In this context, it has been increasingly recognized in recent years that small non-coding RNAs, so-called microRNAs (miRNAs) potentially expand the range of potential biomarkers. These miRNAs are not only present in the cells, but also circulate through the body through the bloodstream and can also be detected in the urine. Previous studies have shown that miRNA profiles in urine differ significantly from healthy women and patients with breast cancer (Erbes et al, 2015). In addition, individual miRNAs could each be assigned specific functions in the cell. Thus, there are numerous studies in which a regulatory effect of certain miRNAs in cell proliferation and consecutive tumor growth could be shown. In addition, important target genes of miRNAs have been identified that de-regulate normal activity in the healthy cell. As a consequence, there is an overexpression or inhibition of genes that ensure the normal cycle of the cell so that cell growth degenerates and the tumor disease takes its course. Although these relationships have been increasingly recognized in recent years, the factors that affect the expression of the miRNAs themselves are not yet understood. These key modulators can provide important information as a biomarker regarding the prognosis of disease progression or treatment response, especially with regard to personalized treatment.
The aim of this project is to investigate the signaling pathways associated with unrestrained cell division leading to the de-regulation of breast cancer-associated miRNAs. Here, the focus lies on investigations in testing of known, proliferation influencing factors such as the receptor-ligand system HER3 and NRG1 and their de-regulating effect on proliferation-relevant miRNAs, such as the miR-205. The identification of the key modulators of miRNAs associated with proliferation provides an innovative approach to use additional biomarkers to predict the disease and predict Therapy response to personalized medicine (keyword: “precision medicine”). In addition, the findings on key regulators of proliferation expand the range of target genes for neoadjuvant therapy concepts.