Our Research
Our research focuses on the biophysics of host-microbe interactions, using and extending quantitative methods to assess how microbes navigate the host environment.
Motivations
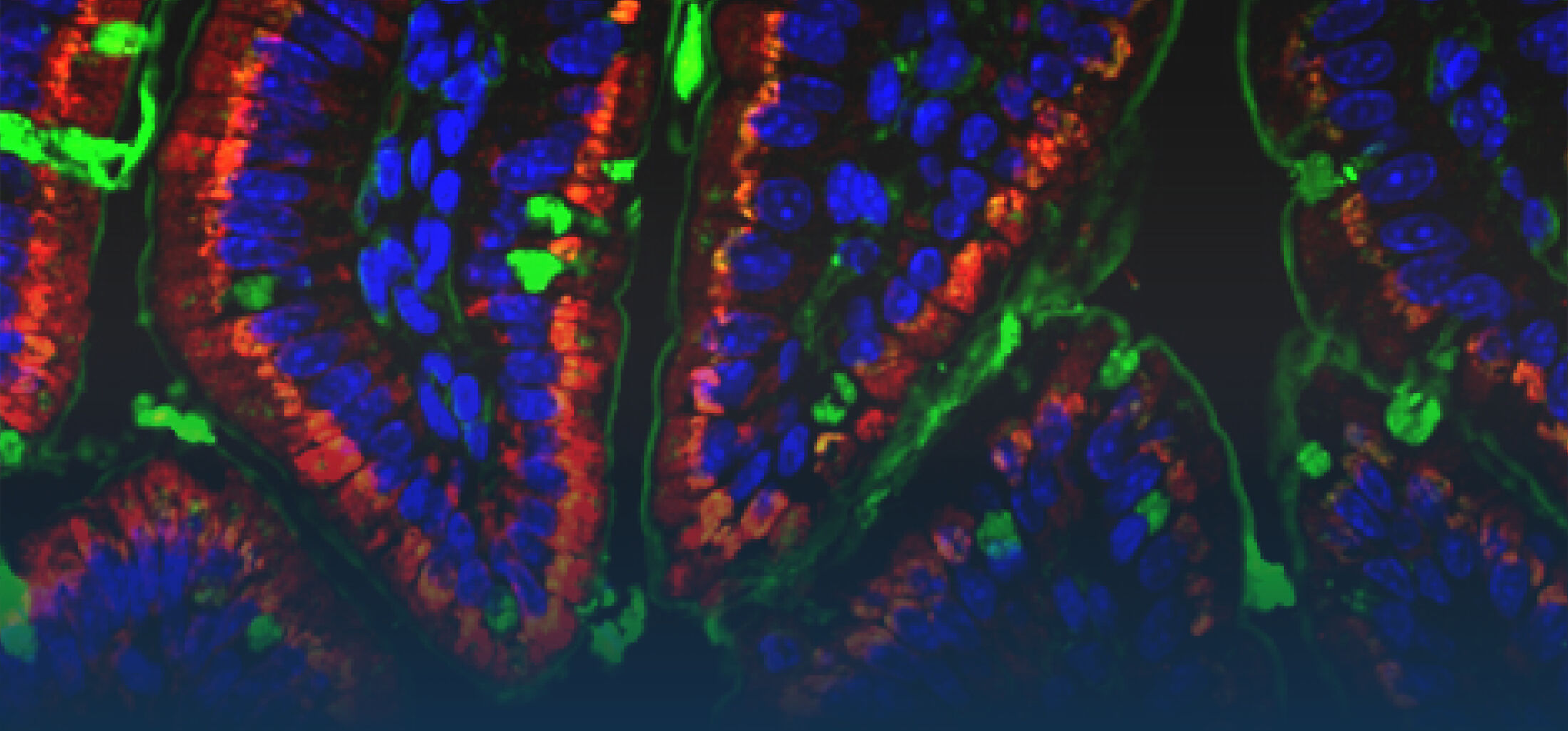
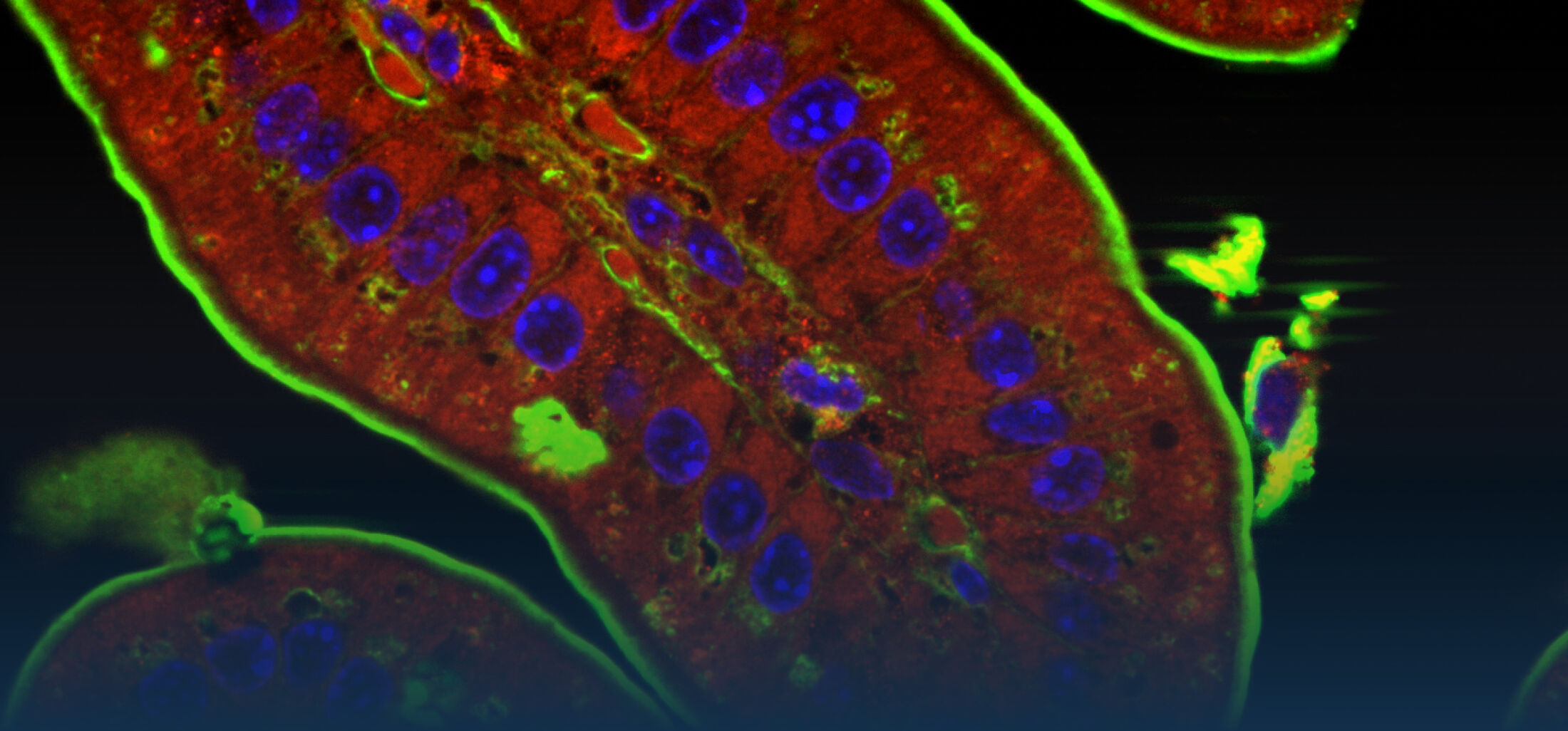
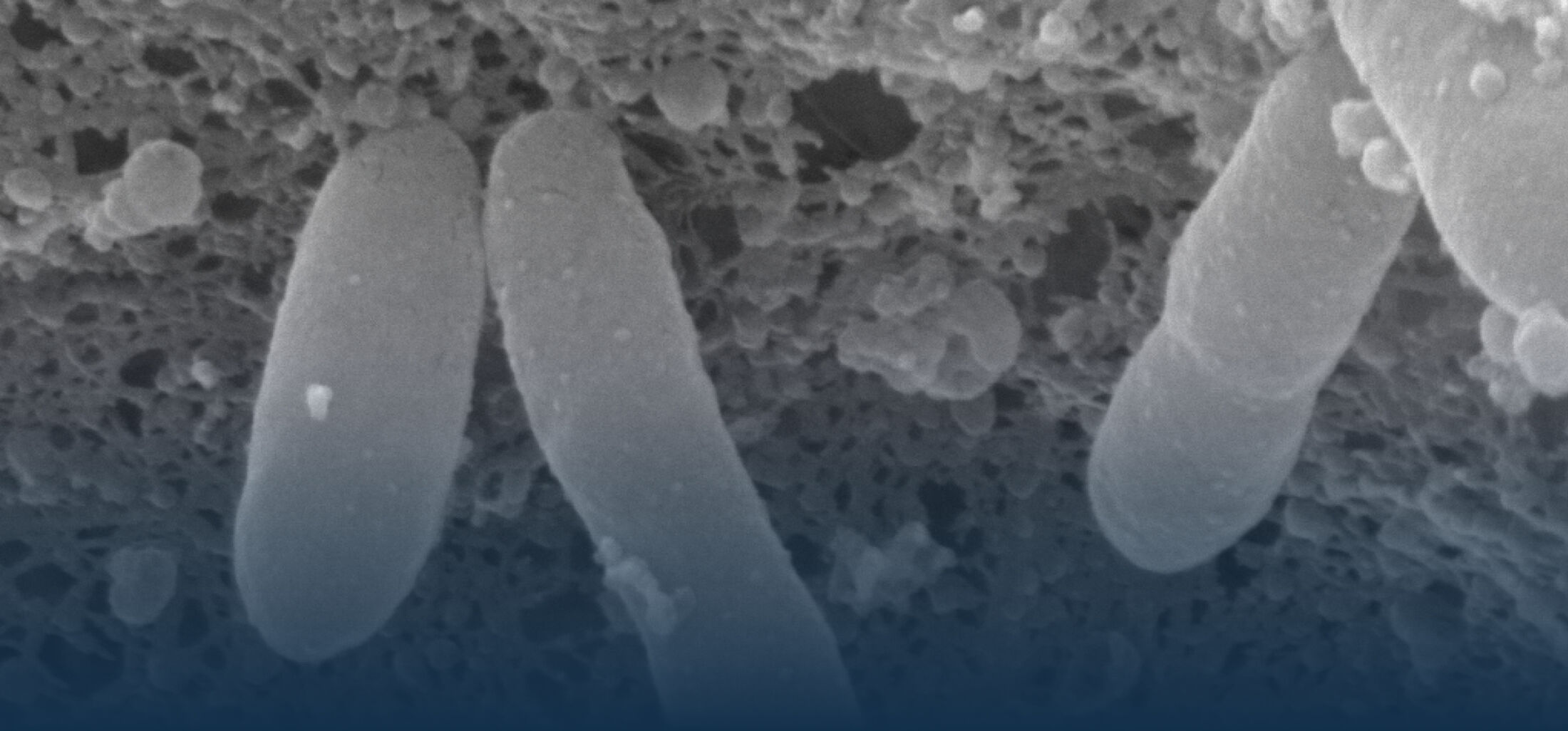
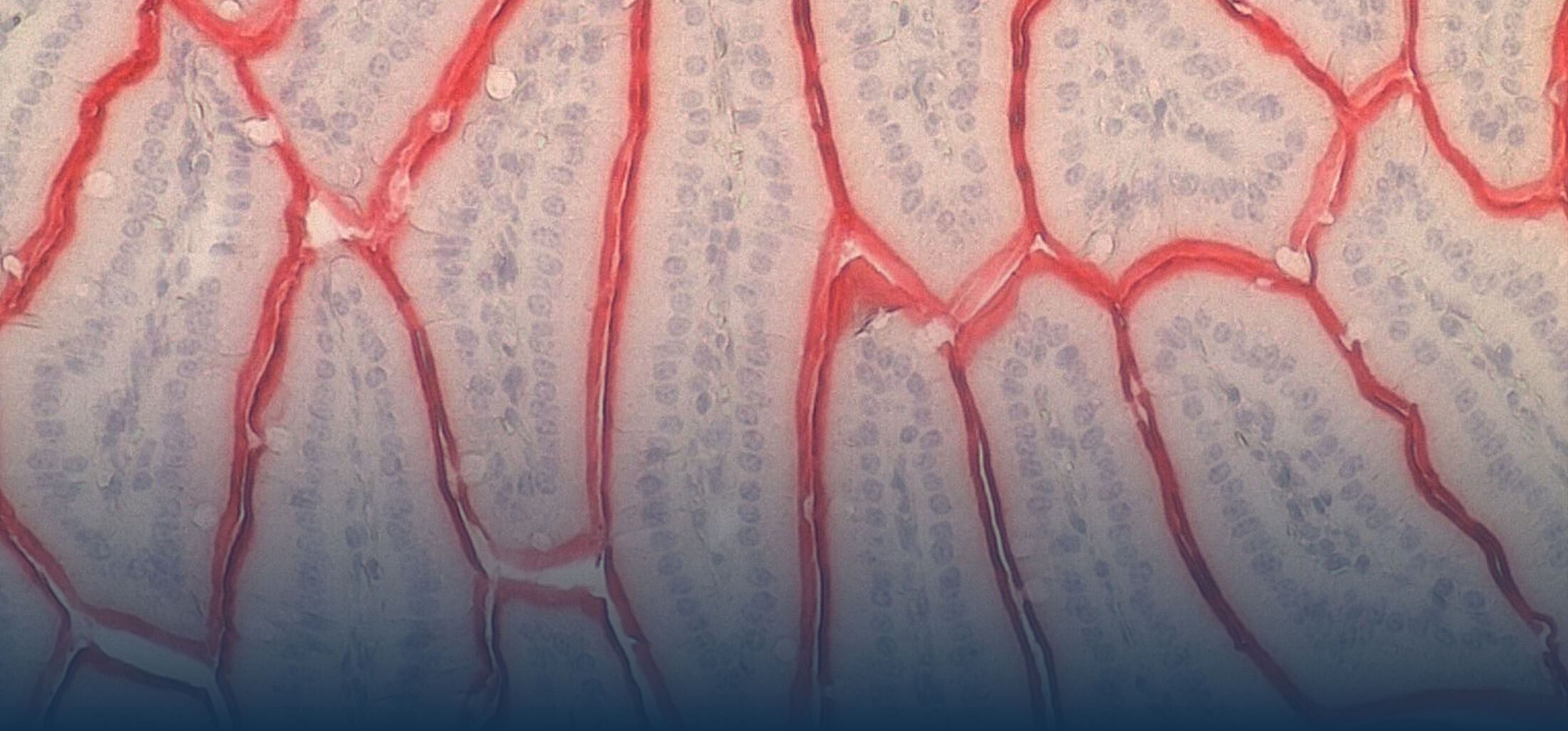
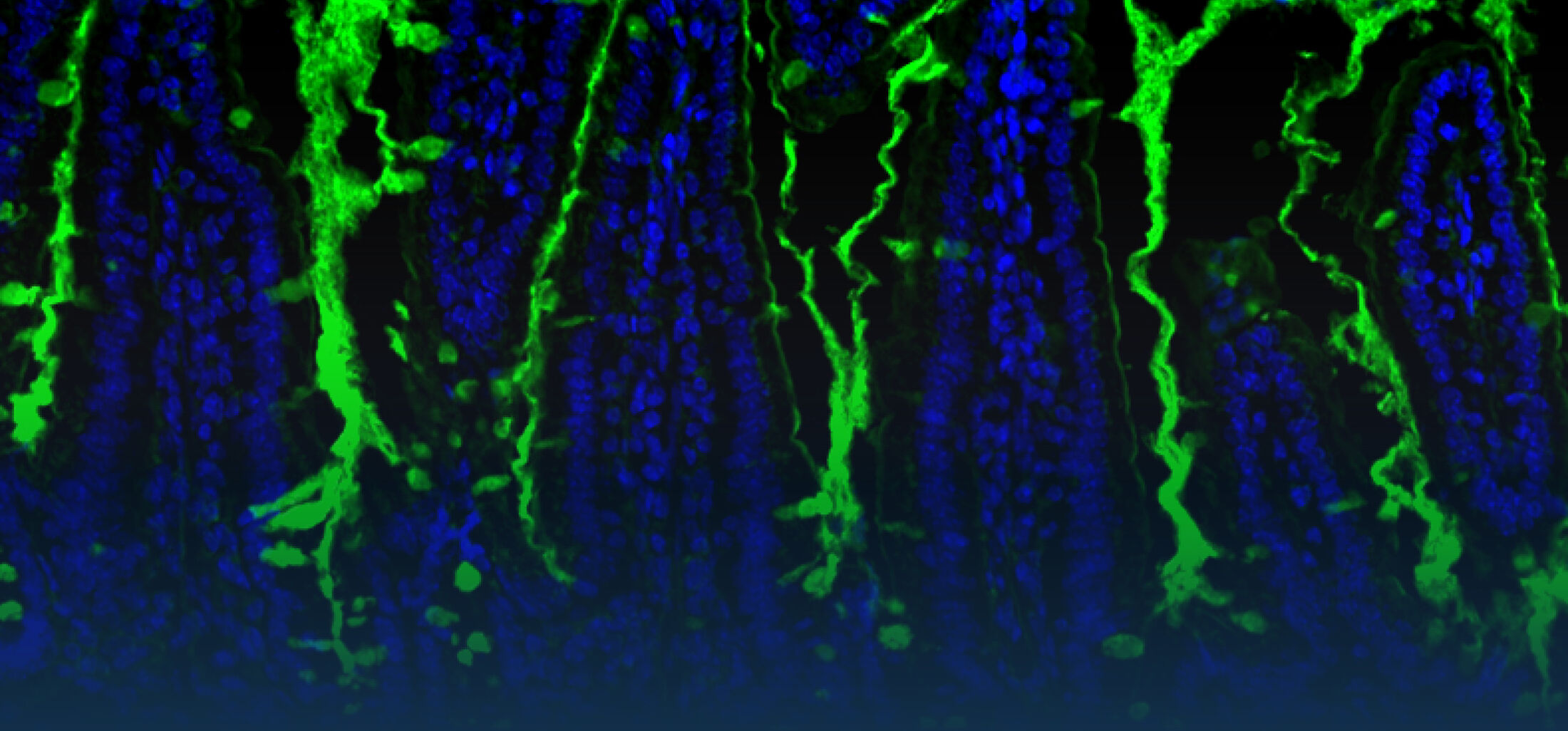
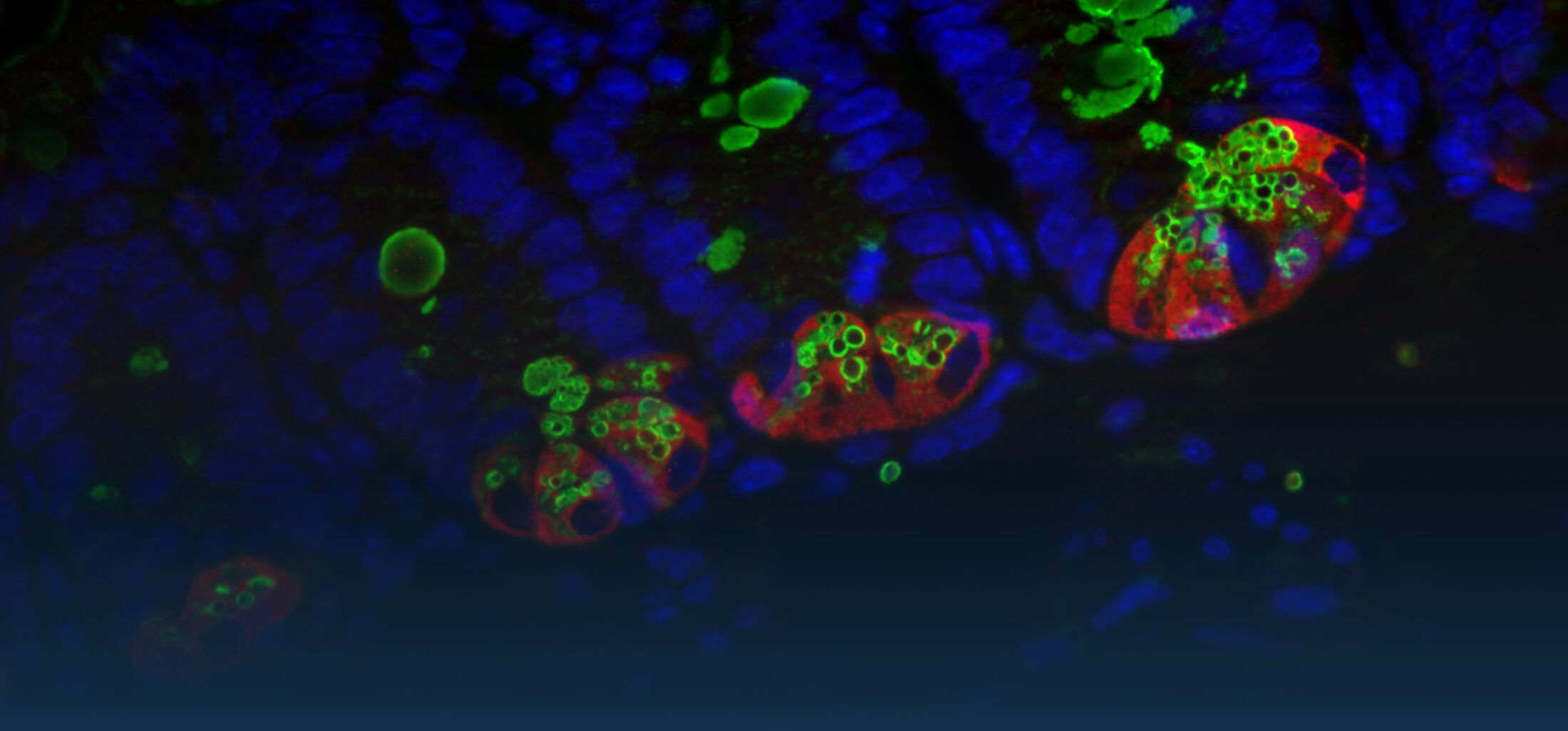
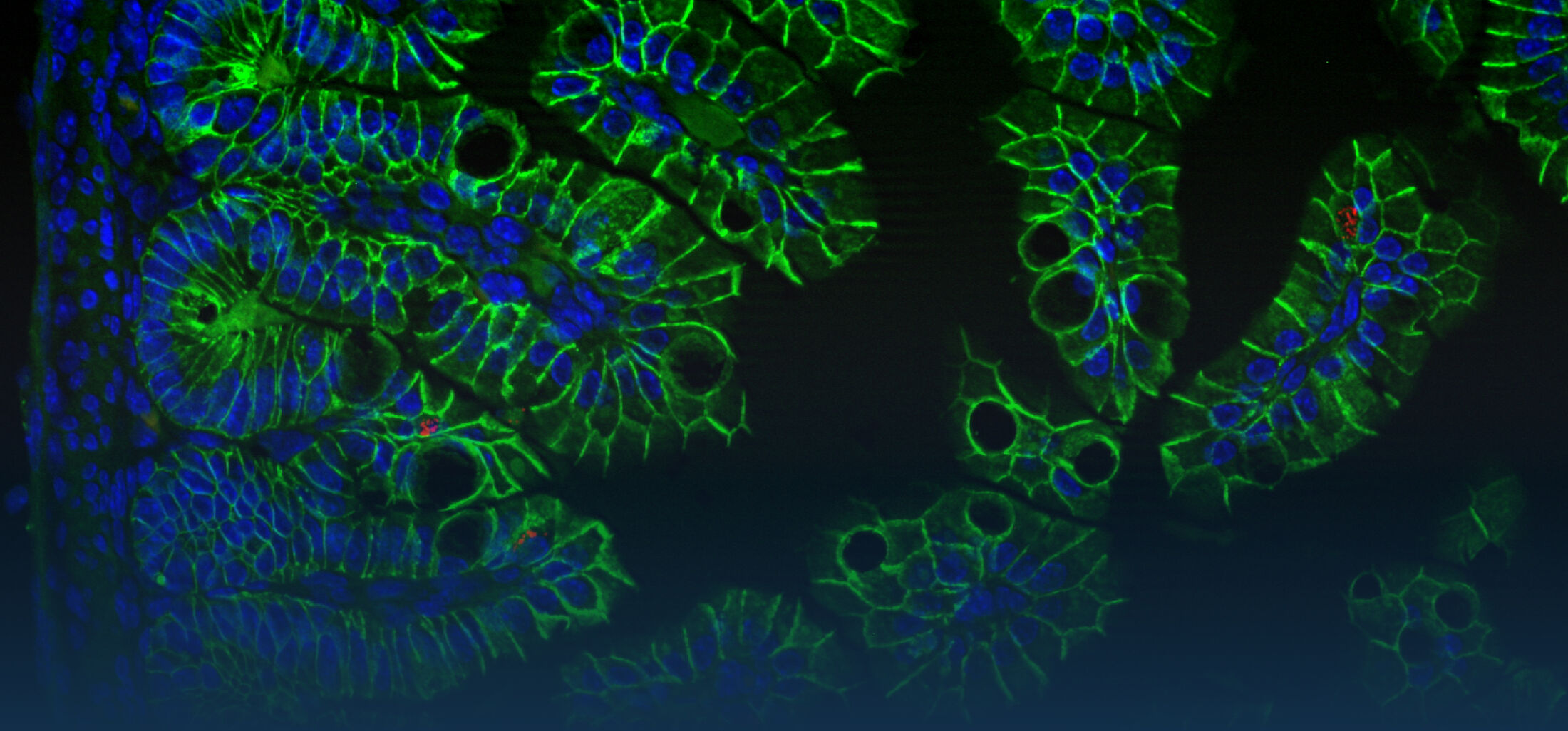
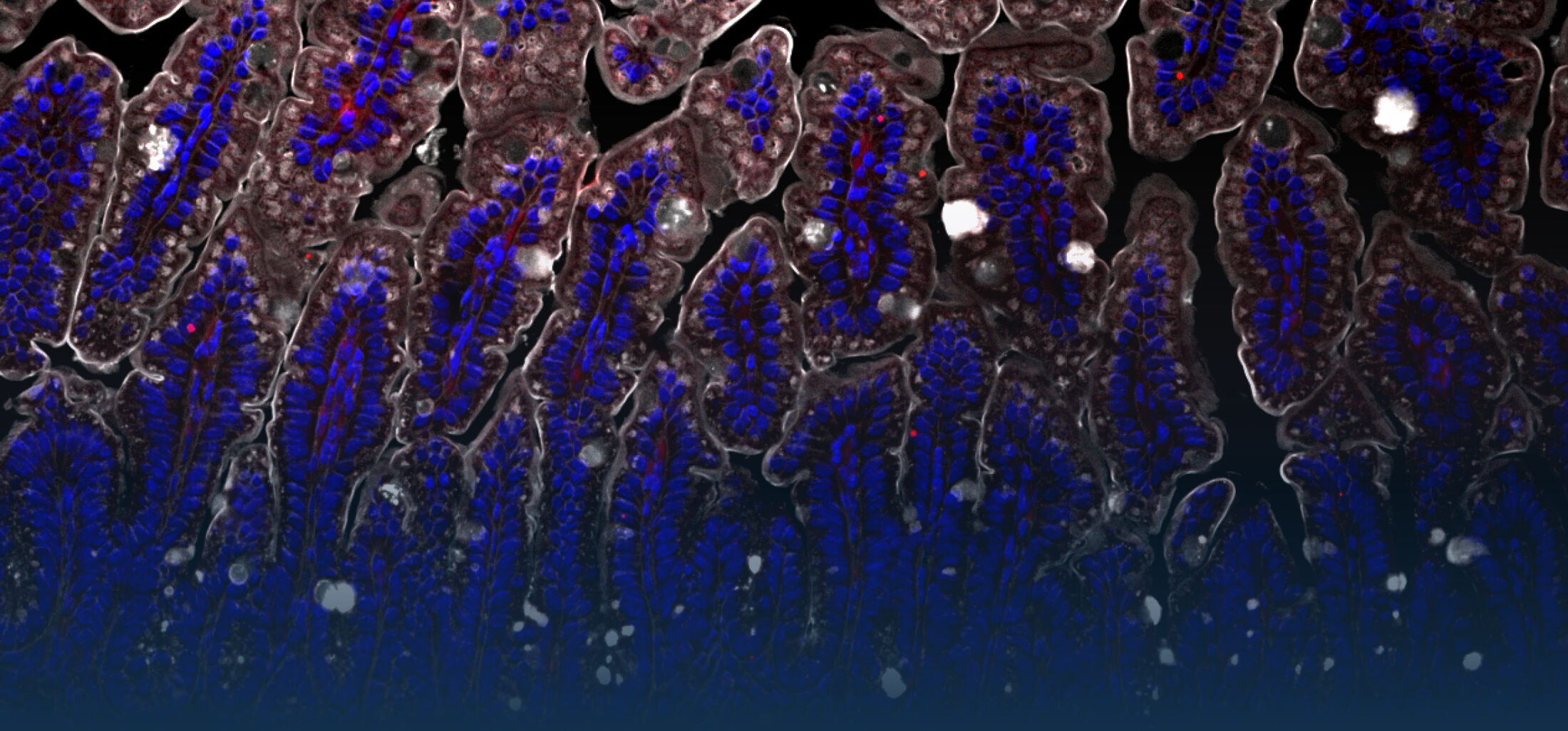
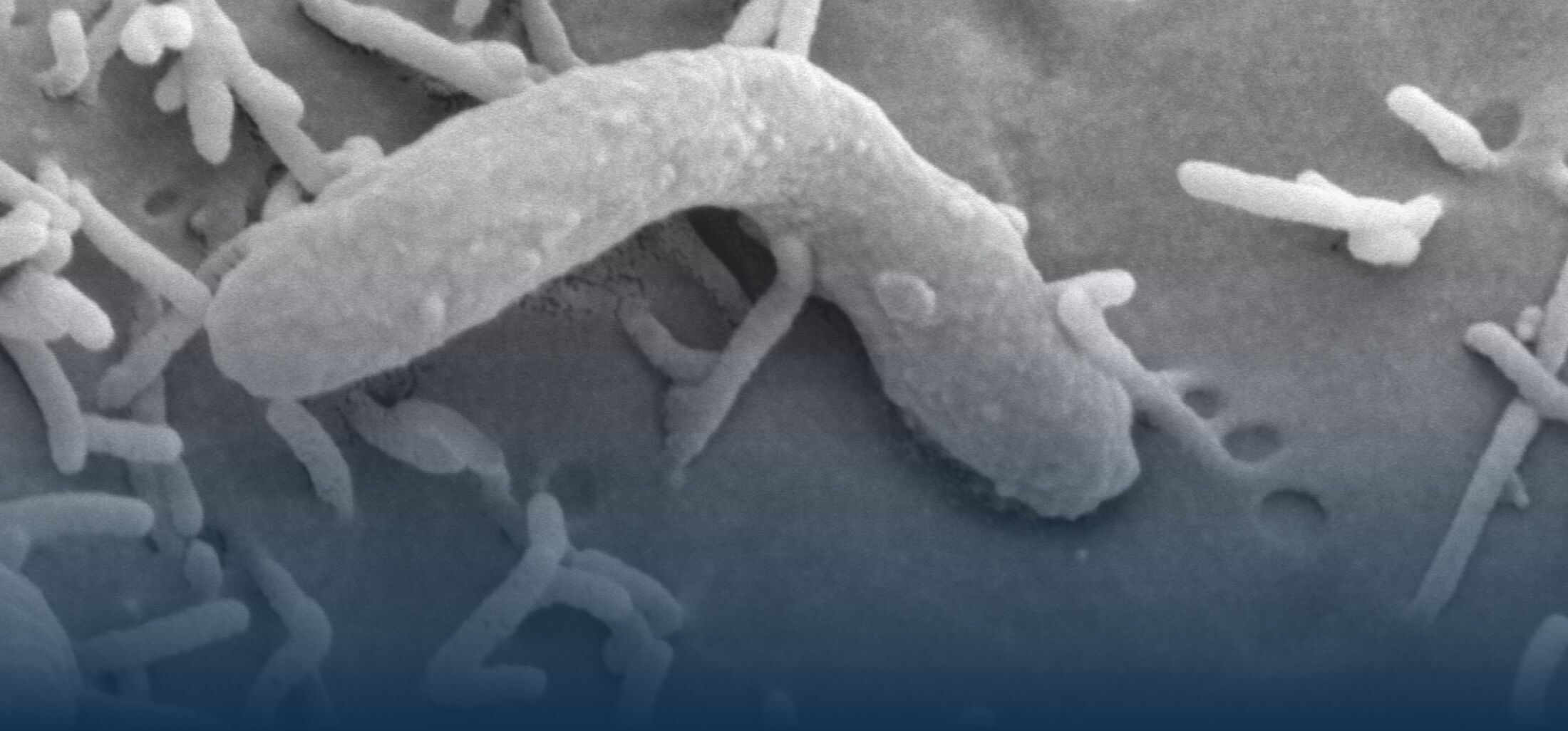
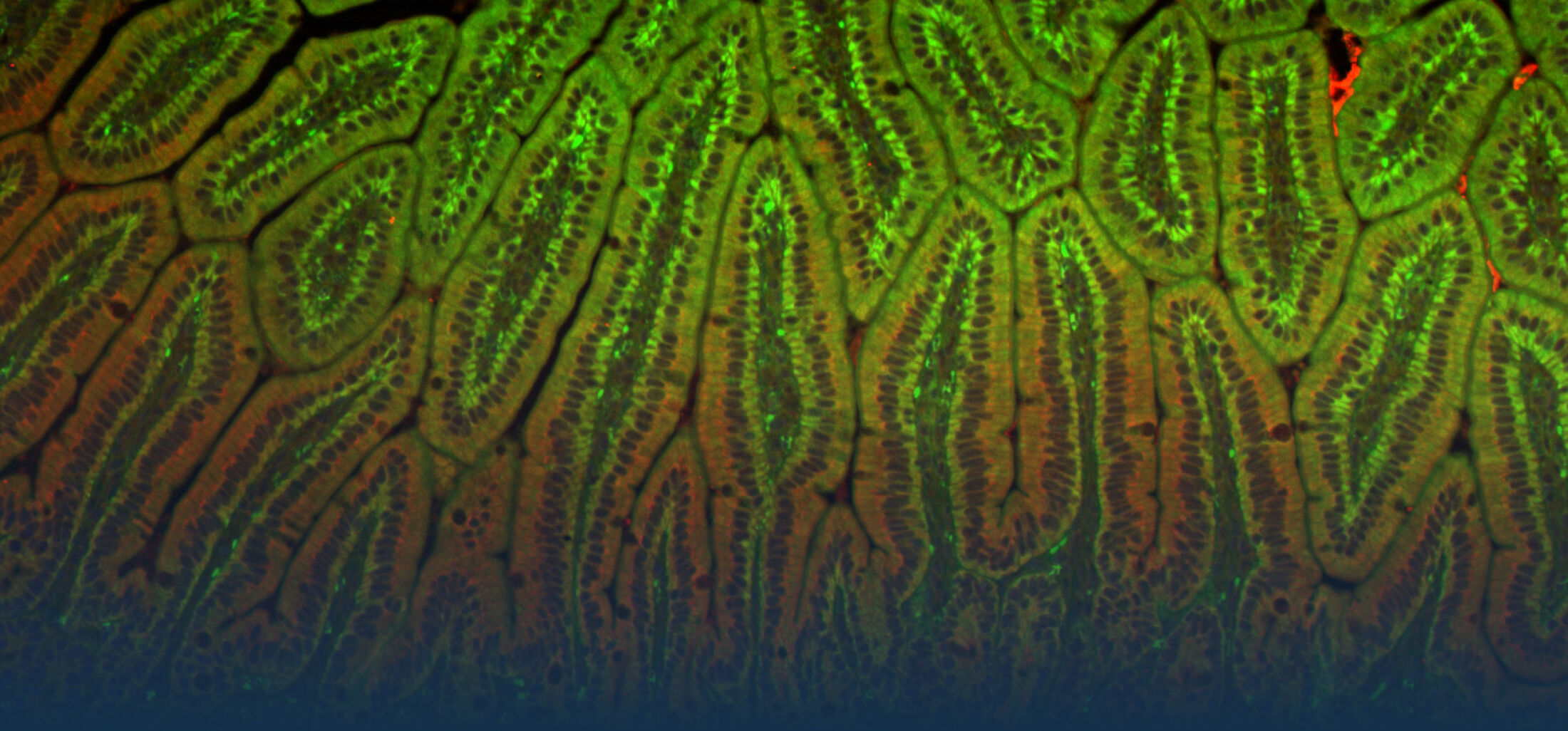
An estimated half of all bacteria can swim by rotating one or more flagella. Most are also capable of biasing this swimming to ascend or descend a chemical gradient in a process called chemotaxis. There are very strong incentives to think that motility and chemotaxis are important in the early steps of inflammation and/or infection, allowing bacteria to breach the mucus barrier that protects our mucous membranes.
Navigation capabilities have been extensively studied in the peritrichously flagellated model organism E. coli in buffer, but much less attention has been paid to:
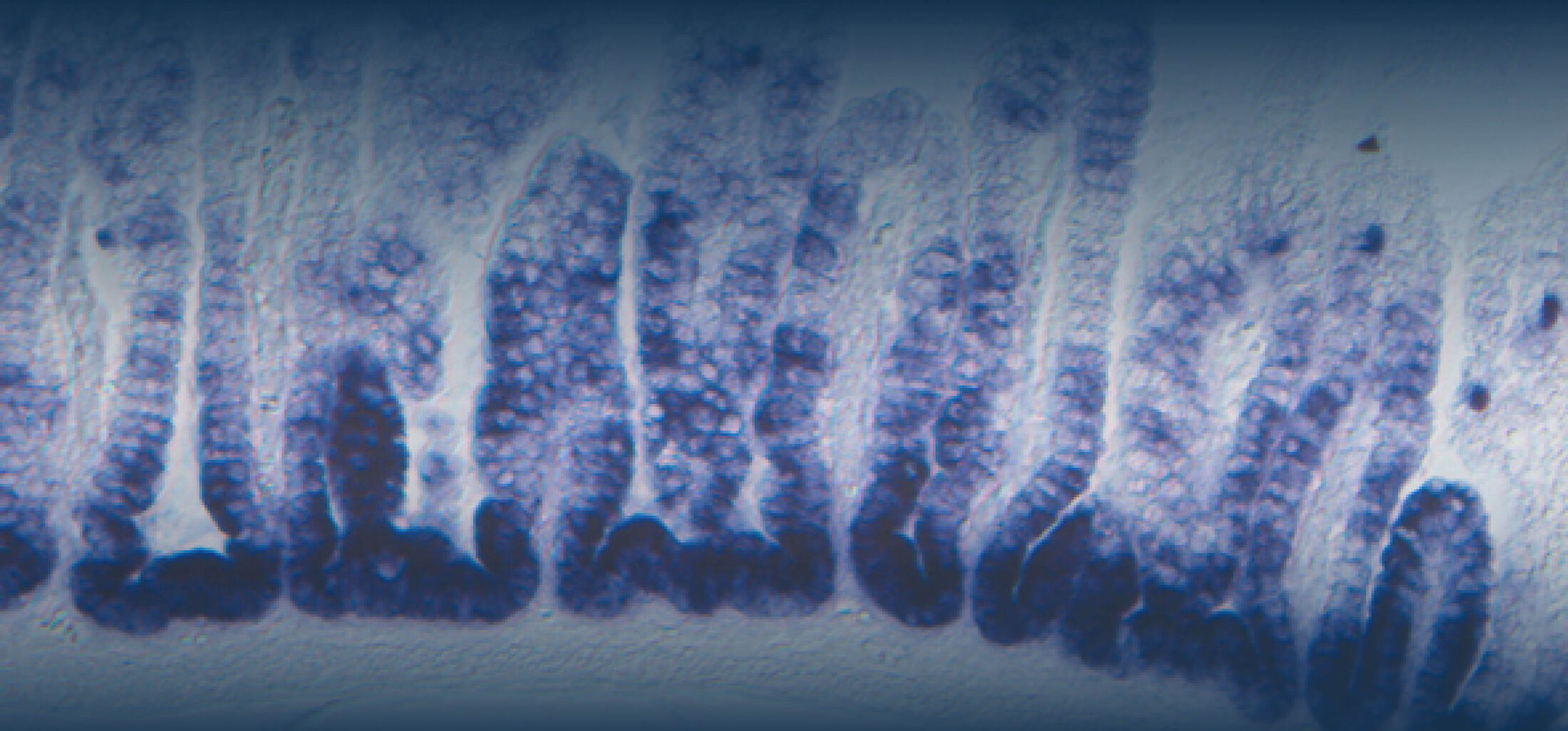
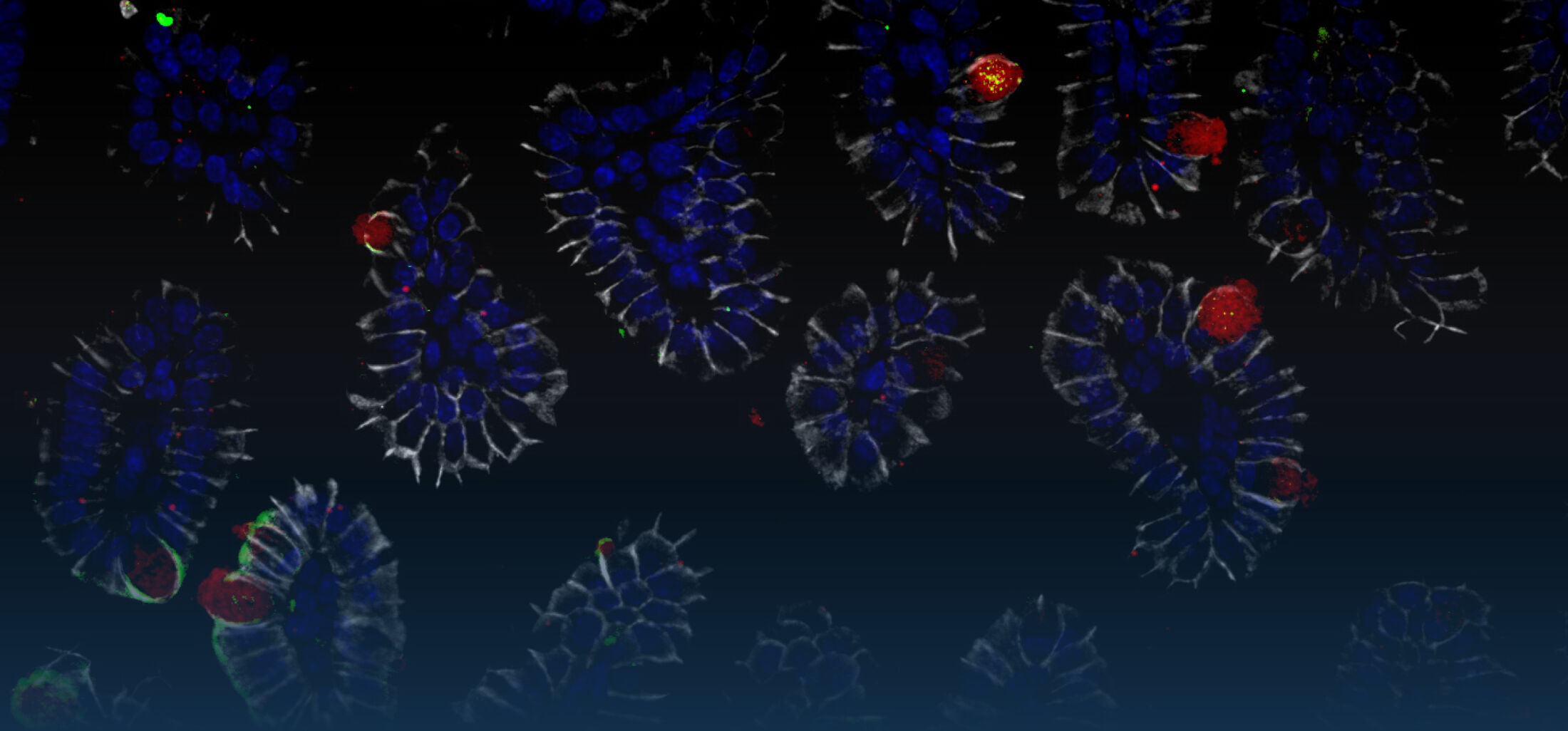
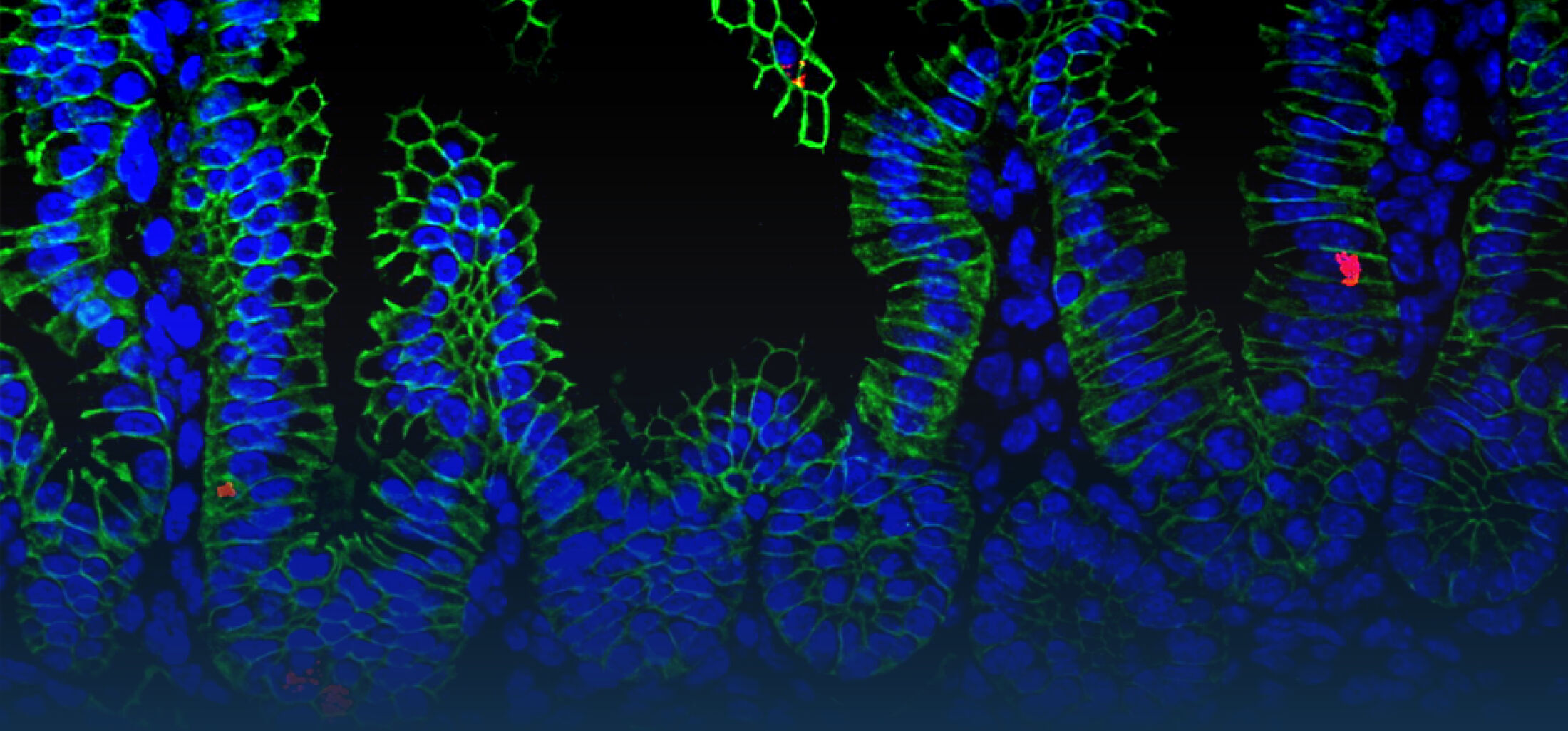
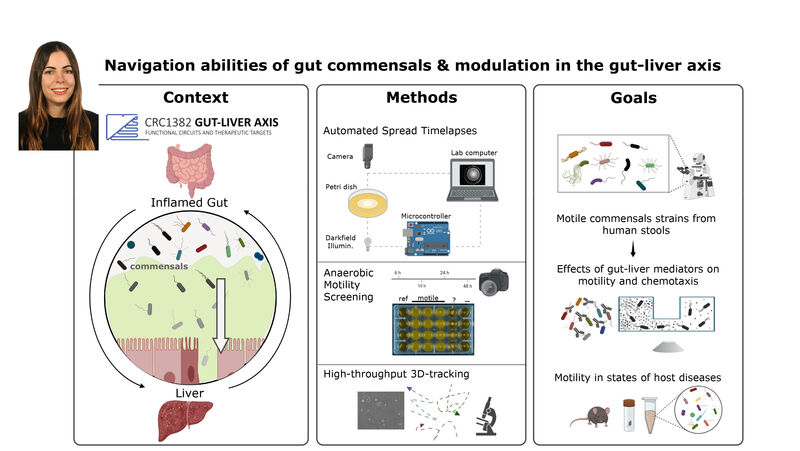
(1) the diversity of motility in other organisms: We ask, for example, how the wide variety of bacterial commensals swim in our gut, and how their motility and chemotaxis may be involved in intestinal inflammatory processes.
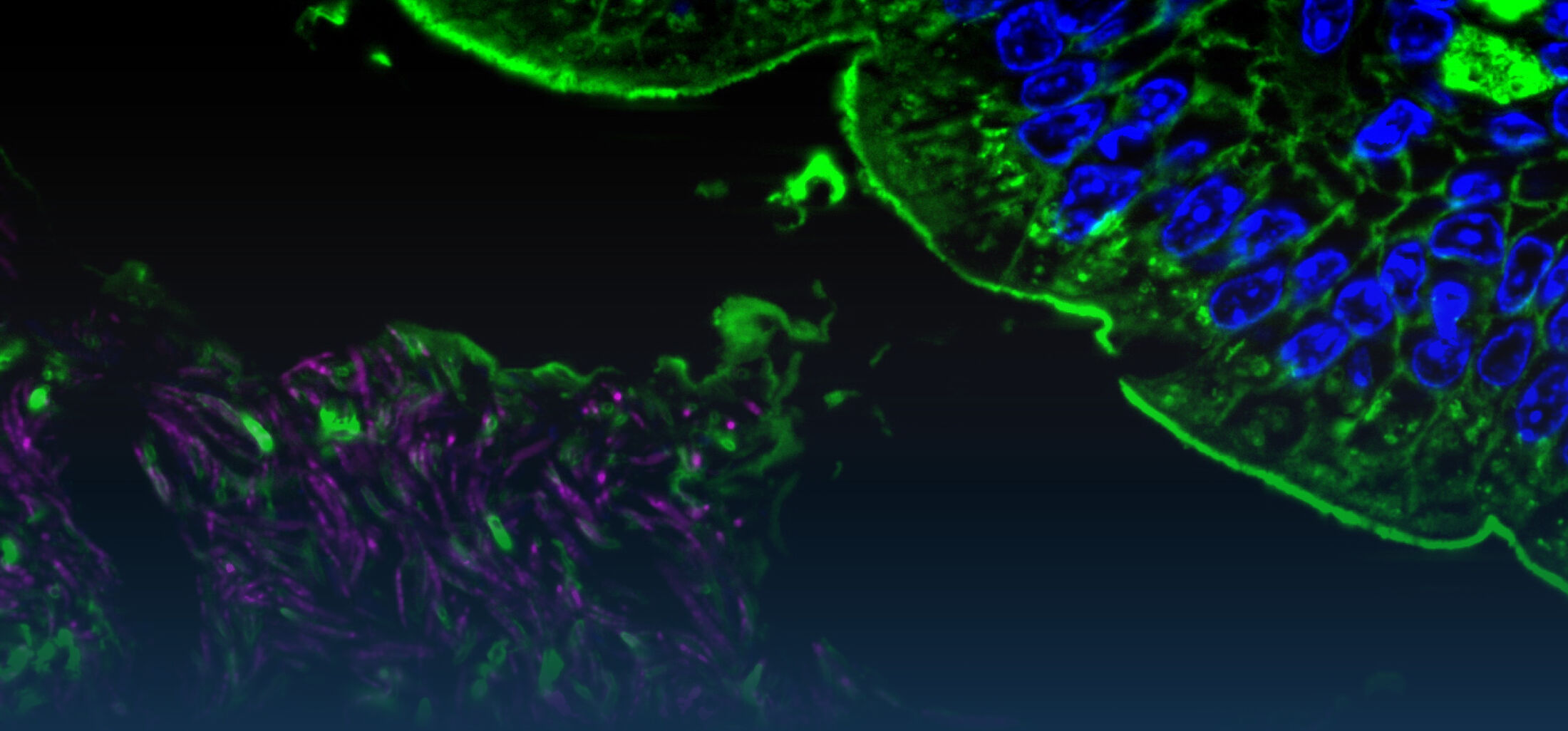
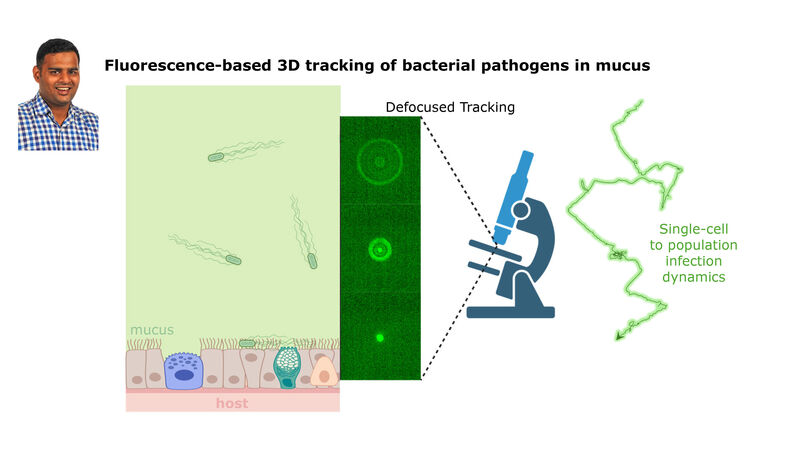
(2) the diversity of motility in viscous or porous environments that more closely mimic the complexity of the host environment: Our current 3D tracking method (see methodological pillars) allows us to track bacteria in buffer, but also in viscous media or agar hydrogels. We are currently developing another bacterial 3D tracking method to assess how pathogens swim in real mucus, from the lumen to the epithelial cell.
Overall we aim to explore the motility and chemotaxis of bacterial pathogens and commensals as a novel and profoundly interdisciplinary angle in the field of host-microbe interactions. In parallel, we are developing simple methods with high species and sample throughputs to screen for triggers or tames of bacterial motility.